Based on TCGA and validation analysis of key lncRNA as a potential prognostic biomarker for gastric cancer patients
-
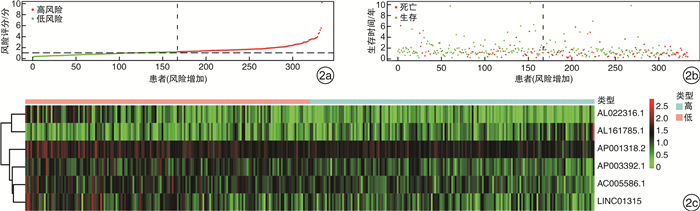
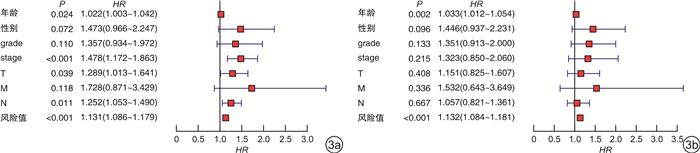
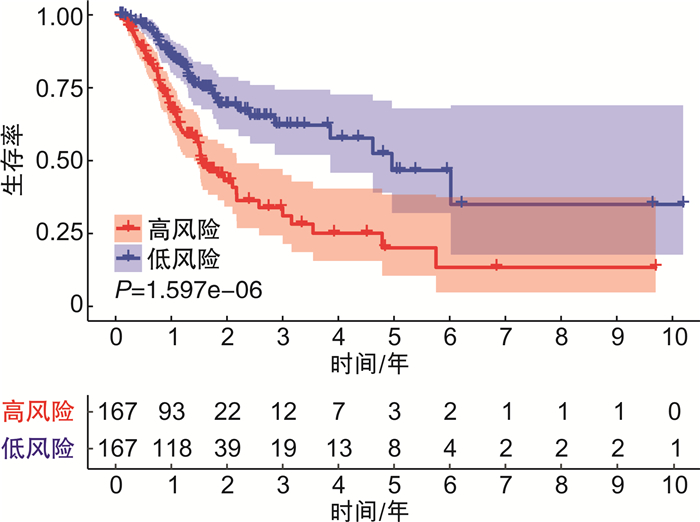
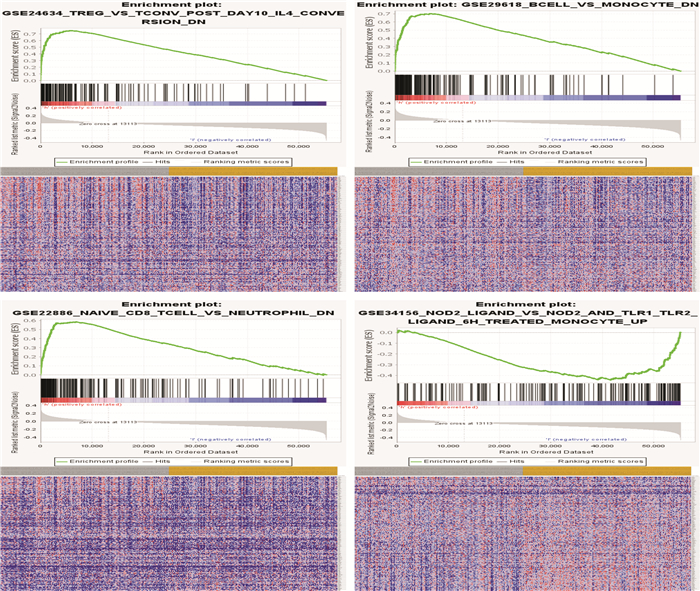
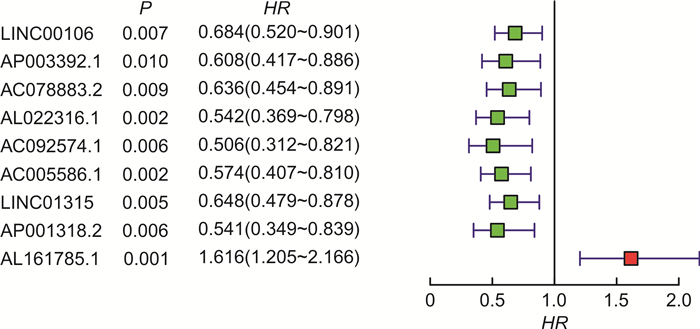
摘要: 目的 构建基于长链非编码RNA(lncRNA)的胃癌(GCa)患者预后风险模型。方法 从癌症基因组图谱(TCGA)数据库中检索GCa患者的RNA测序数据和临床信息,使用R软件筛选免疫相关基因及lncRNA,进行单变量和多变量Cox回归分析以构建预后风险模型、生存分析和受试者ROC曲线用于评估模型的敏感性和特异性。结果 从TCGA数据库收集的GCa患者的临床信息,以确定用于预后的lncRNA生物标志物。从375例胃腺癌(stomach adenocarcinoma,STAD)组织和32例非癌组织中根据单变量和多变量Cox回归分析,发现了6个lncRNA(AP003392.1、AL022316.1、AC005586.1、LINC01315、AP001318.2、AL161785.1)与GCa患者存活相关,被选择建立预后风险模型。基于该风险模型,高危患者和低危患者的总生存率存在显著差异。该模型的ROC曲线下面积(AUC)为0.686。基因表达谱数据动态分析(gene expression profiling interactive analysis,GEPIA)结果证实了LINC01315在STAD中的表达和预后意义。临床数据证实胃癌中LINC01315的表达较健康人群是高表达。结论 构建lncRNA预后风险模型,风险值可以作为独立预后因子预测GCa患者的预后情况,降低LINC01315表达可能在GCa患者的生存率方面发挥重要作用。
-
关键词:
- 长链非编码RNA /
- 癌症基因组图谱 /
- 基因表达谱数据动态分析 /
- 预后风险模型
Abstract: Objective To construct a lncRNA-based prognostic risk model for patients with gastric cancer(GCa).Methods RNA sequencing data and clinical information of GCa patients were retrieved from The Cancer Genome Atlas(TCGA) database, immune-related genes and lncRNAs were screened using R software, and univariate and multivariate Cox regression analyses were performed to construct prognostic risk models, survival analyses and subject ROC curves for assessing the sensitivity and specificity of the models.Results Collected clinical information on GCa patients from the TCGA database to identify lncRNA biomarkers for prognostic purposes. From 375 stomach adenocarcinoma(STAD) tissues and 32 non-cancerous tissues based on univariate and multivariate Cox regression analysis, six lncRNAs(AP003392.1, AL022316.1, AC005586.1, LINC01315, AP001318.2, and AL161785.1) were associated with survival in GCa patients and were selected for prognostic risk modeling. Based on this risk model, there was a significant difference in overall survival between high-risk and low-risk patients. The area under the curve(AUC) of the ROC curve for this model was 0.686. Gene Expression Profiling Interactive Analysis(GEPIA) results confirmed the expression and prognostic significance of LINC01315 in STAD. The clinical data confirmed that the expression of LINC01315 in STAD is high compared to the healthy population.Conclusion LncRNA prognostic risk model was constructed, and the risk value could be used as an independent prognostic factor to predict the prognosis of GCa patients, and reducing LINC01315 expression may play an important role in the survival of GCa patients. -
-
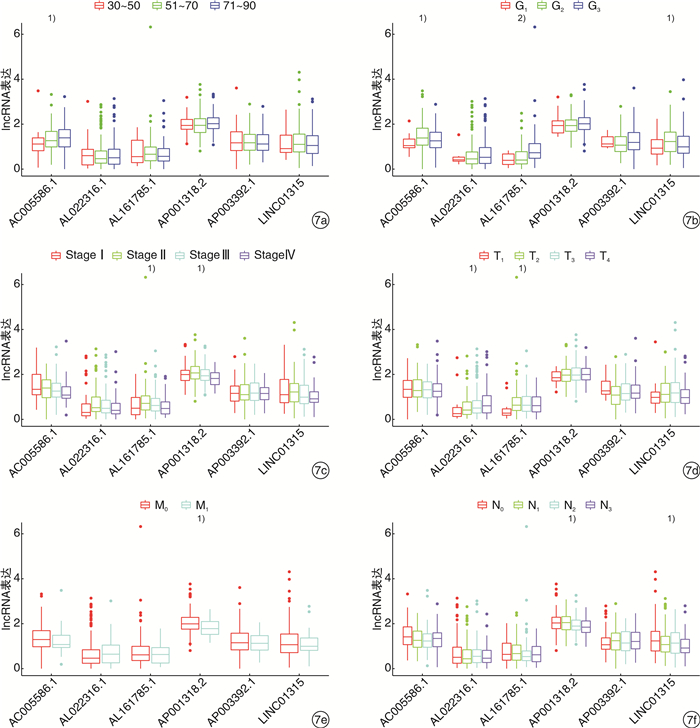
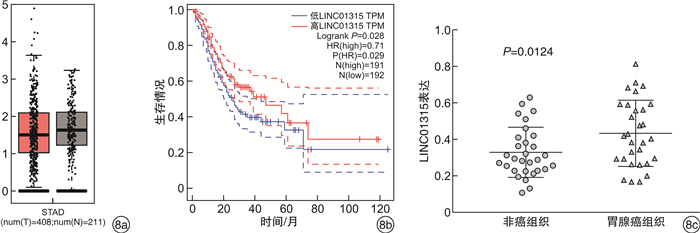
表 1 免疫lncRNA模型
id coef HR HR.95L HR.95H P AP003392.1 -0.62778 0.533777 0.359633 0.792247 0.001834 AL022316.1 -0.55926 0.571631 0.37932 0.86144 0.007523 AC005586.1 -0.36064 0.697229 0.481281 1.010072 0.056524 LINC01315 -0.27145 0.762272 0.548309 1.059728 0.106343 AP001318.2 -0.39167 0.675929 0.418855 1.090782 0.108695 AL161785.1 0.591984 1.807571 1.37336 2.379065 2.41e-5 表 2 LINC01315与胃腺癌患者临床特征的相关性
例 临床特征 LINC01315 低表达 高表达 总例数 P 年龄/岁 0.273 >45 8 12 20 < 45 2 8 10 性别 0.192 男 11 13 24 女 1 5 6 肿瘤大小 0.044 T1~T2 7 14 21 T3~T4 6 3 9 淋巴结状况 0.770 N0 7 15 22 N1 3 5 8 TNM分期 0.765 Ⅰ~Ⅱ 8 10 18 Ⅲ~Ⅳ 6 6 12 -
[1] Li C, Wang H, Meng S, et al. lncRNA GAS8-AS1 regulates cancer cell proliferation and predicts poor survival of patients with gastric cancer[J]. Oncol Lett, 2022, 23(2): 48.
[2] Xu Z, Ran J, Gong K, et al. LncRNA SUMO1P3 regulates the invasion, migration and cell cycle of gastric cancer cells through Wnt/β-catenin signaling pathway[J]. J Recept Signal Transduct Res, 2021, 41(6): 574-581. doi: 10.1080/10799893.2020.1836494
[3] Yoshida T, Yamaguchi T, Maekawa S, et al. Identification of early genetic changes in well-differentiated intramucosal gastric carcinoma by target deep sequencing[J]. Gastric Cancer, 2019, 22(4): 742-750. doi: 10.1007/s10120-019-00926-y
[4] Andergassen D, Rinn JL. From genotype to phenotype: genetics of mammalian long non-coding RNAs in vivo[J]. Nat Rev Genet, 2022, 23(4): 229-243.
[5] Hu X, Sood AK, Dang CV, et al. The role of long noncoding RNAs in cancer: the dark matter matters[J]. Curr Opin Genet Dev, 2018, 48: 8-15. doi: 10.1016/j.gde.2017.10.004
[6] Chen S. LINC00852 Regulates Cell Proliferation, Invasion, Migration and Apoptosis in Hepatocellular Carcinoma Via the miR-625/E2F1 Axis[J]. Cell Mol Bioeng, 2021, 15(2): 207-217.
[7] Luo J, Xie K, Gao X, et al. Long Noncoding RNA Nuclear Paraspeckle Assembly Transcript 1 Promotes Progression and Angiogenesis of Esophageal Squamous Cell Carcinoma Through miR-590-3p/MDM2 Axis[J]. Front Oncol, 2020, 10: 618930.
[8] Lin X, Zhuang S, Chen X, et al. lncRNA ITGB8-AS1 functions as a ceRNA to promote colorectal cancer growth and migration through integrin-mediated focal adhesion signaling[J]. Mol Ther, 2022, 30(2): 688-702. doi: 10.1016/j.ymthe.2021.08.011
[9] Chen J, Liao X, Cheng J, et al. Targeted Methylation of the LncRNA NEAT1 Suppresses Malignancy of Renal Cell Carcinoma[J]. Front Cell Dev Biol, 2021, 9: 777349. doi: 10.3389/fcell.2021.777349
[10] Gao L, Xue J, Liu X, et al. A risk model based on autophagy-related lncRNAs for predicting prognosis and efficacy of immunotherapy and chemotherapy in gastric cancer patients[J]. Aging(Albany NY), 2021, 13(23): 25453-25465.
[11] Hao H, Chen H, Xie L, et al. LncRNA KCNQ1OT1 Promotes Proliferation, Invasion and Metastasis of Prostate Cancer by Regulating miR-211-5p/CHI3 L1 Pathway[J]. Onco Targets Ther, 2021, 14: 1659-1671. doi: 10.2147/OTT.S288785
[12] Wang FH, Zhang XT, Li YF, et al. The Chinese Society of Clinical Oncology(CSCO): Clinical guidelines for the diagnosis and treatment of gastric cancer, 2021[J]. Cancer Commun(Lond), 2021, 41(8): 747-795.
[13] Lyons K, Le LC, Pham YT, et al. Gastric cancer: epidemiology, biology, and prevention: a mini review[J]. Eur Cancer Prev, 2019, 28(5): 397-412. doi: 10.1097/CEJ.0000000000000480
[14] Necula L, Matei L, Dragu D, et al. Recent advances in gastric cancer early diagnosis[J]. World J Gastroenterol, 2019, 25(17): 2029-2044. doi: 10.3748/wjg.v25.i17.2029
[15] Zhuo W, Liu Y, Li S, et al. Long Noncoding RNA GMAN, Up-regulated in Gastric Cancer Tissues, Is Associated With Metastasis in Patients and Promotes Translation of Ephrin A1 by Competitively Binding GMAN-AS[J]. Gastroenterology, 2019, 156(3): 676-691. e11. doi: 10.1053/j.gastro.2018.10.054
[16] Zhang E, He X, Zhang C, et al. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1[J]. Genome Biol, 2018, 19(1): 154. doi: 10.1186/s13059-018-1523-0
[17] Liang R, Zhang J, Zhang RM, et al. LINC01315 silencing inhibits the aggressive phenotypes of colorectal carcinoma by sponging miR-205-3p[J]. Biochem Biophys Res Commun, 2021, 534: 1033-1039. doi: 10.1016/j.bbrc.2020.10.041
[18] Chen FB, Wu P, Zhou R, et al. LINC01315 Impairs microRNA-211-Dependent DLG3 Downregulation to Inhibit the Development of Oral Squamous Cell Carcinoma[J]. Front Oncol, 2020, 10: 556084. doi: 10.3389/fonc.2020.556084
[19] Ren J, Zhang FJ, Wang JH, et al. LINC01315 promotes the aggressive phenotypes of papillary thyroid cancer cells by sponging miR-497-5p[J]. Kaohsiung J Med Sci, 2021, 37(6): 459-467. doi: 10.1002/kjm2.12369
[20] Gao W, Chan JY, Wong TS. Differential expression of long noncoding RNA in primary and recurrent nasopharyngeal carcinoma[J]. Biomed Res Int, 2014, 2014: 404567.
[21] Naorem LD, Prakash VS, Muthaiyan M, et al. Comprehensive analysis of dysregulated lncRNAs and their competing endogenous RNA network in triple-negative breast cancer[J]. Int J Biol Macromol, 2020, 145: 429-436. doi: 10.1016/j.ijbiomac.2019.12.196
-



 下载:
下载: